export LINBUGS_HOME="/c/tduong/home/bin/openbugs" export LINBUGS_TEMP="/c/tduong/home/temp"where LINBUGS_HOME is the directory where you want to install LinBugs and LINBUGS_TEMP is the directory where LinBugs writes its temporary files.
http://mathstat.helsinki.fi/openbugsUnpack this into $LINBUGS_HOME. After unpacking, check that there is a file called cbugs. If not, download it from
http://www.gatsby.ucl.ac.uk/~iam23/compnotes/openbugsIf there is already a file called linbugs in $LINBUGS_HOME, copy it to another file e.g. linbugs.orig. Create a file called linbugs containing the following commands
#!/bin/bash
export LD_ASSUME_KERNEL=2.4.1
cd $LINBUGS_HOME
if [ \! -e $LINBUGS_TEMP ] ; then
mkdir $LINBUGS_TEMP
fi
if [ -e bugs.so ] ; then
./cbugs "$LINBUGS_HOME" "$LINBUGS_TEMP" "/bugs.so"
else
./cbugs "$LINBUGS_HOME" "$LINBUGS_TEMP" "/brugs.so"
fi
./linbugsSomething like
OpenBUGS ClassicBUGS release 2.1.1 type 'modelQuit()' to quit Bugs>should appear in the command line. Quit from LinBugs by typing modelQuit().
#########################################################################
## Simple R interface for LinBugs
#########################################################################
##########################################################################
## Execute LinBugs from inside R session
##
## Parameters
## file - file of LinBUGS commands
##
## Returns
## Invisible
##########################################################################
rlinbugs.exec <- function(file)
{
system(paste("bash $LINBUGS_HOME/linbugs <", file), ignore.stderr=TRUE)
invisible()
}
##########################################################################
## Reads CODA output files into R objects
##
## Parameters
## stem - stem for CODA output files
## chain.num - chain number
##
## Returns
## Matrix with 3 columns
## first column is "iter" = iteration number
## subsequent columns are the values of the each iteration of parameters
## in MCMC
#########################################################################
rlinbugs.read <- function(stem, chain.num=1)
{
varnames <- read.table(paste(stem,"CODAindex.txt", sep=""))
n.var <- nrow(varnames)
n.iter <- varnames[1,3]
output <- matrix(0, nrow=n.iter, ncol=(n.var+1))
for (i in 1:n.var)
{
if (i==1)
{
output.temp <- read.table(paste(stem,"CODAchain", chain.num, ".txt", sep=""),
nrows=n.iter)
output[,1:2] <- as.matrix(output.temp)
}
else if (i>1)
{
output.temp <- read.table(paste(stem,"CODAchain", chain.num, ".txt", sep=""),
skip=(i-1)*n.iter, nrows=n.iter)
output[,i+1] <- as.matrix(output.temp)[,2]
}
}
colnames(output) <- c("iter", as.character(varnames[,1]))
return(output)
}
Save these into rlinbugs.R in your working directory. In my case it is,
/c/tduong/home/research/bugs.
price = a + b*age
In your working directory, create the following files. Unfortunately I can't quite force LinBUGS to read in files from the working directory (its default is to use $LINBUGS_HOME), so you have to specify the full address of all input/ouput files.
| File name | Commands |
|---|---|
| carsbugs.txt |
modelCheck("/c/tduong/home/research/bugs/carsmodel.txt")
modelData("/c/tduong/home/research/bugs/carsdata.txt")
modelCompile()
modelInits("/c/tduong/home/research/bugs/carsinits.txt")
modelUpdate(10000)
summarySet("a")
summarySet("b")
samplesSet("a")
samplesSet("b")
modelUpdate(10000)
summaryStats("*")
samplesStats("*")
samplesCoda("*","/c/tduong/home/research/bugs/output")
modelQuit()
|
| carsmodel.txt |
model
{
for( i in 1 : N ) {
mu[i] <- a+b*age[i]
price[i] ~ dnorm(mu[i], tau)
}
tau ~ dgamma(0.001, 0.001)
sigma <- 1 / sqrt(tau)
a ~ dnorm(0, 1.0E-12)
b ~ dnorm(0, 1.0E-12)
}
|
| carsinits.txt |
list( a=0, b=0, tau=1) |
| carsdata.txt |
list(N=39, age = c(13, 14, 14,12, 9, 15, 10, 14, 9, 14, 13, 12, 9, 10, 15, 11, 15, 11, 7, 13, 13, 10, 9, 6, 11, 15, 13, 10, 9, 9, 15, 14, 14, 10, 14, 11, 13, 14, 10), price = c(2950, 2300, 3900, 2800, 5000, 2999, 3950, 2995, 4500, 2800, 1990, 3500, 5100, 3900, 2900, 4950, 2000, 3400, 8999, 4000, 2950, 3250, 3950, 4600, 4500, 1600, 3900, 4200, 6500, 3500, 2999, 2600, 3250, 2500, 2400, 3990, 4600, 450, 4700)) |
In the samplesCoda command in carsbugs.txt, the second argument is known as the stem, and it creates output files of the form outputCODAchainN.txt where N is the chain number, and outputCODAindex.txt, in the directory /c/tduong/home/research/bugs
Note that graphical functions are not yet (as of 31 Dec 2005) available in LinBugs.
source("rlinbugs.R")
rlinbugs.exec(file="carsbugs.txt")
cars <- rlinbugs.read(stem="/c/tduong/home/research/bugs/output", chain.num=1)
The argument to stem in rlinbugs.read
should be exactly the same as the stem in the
samplesCoda.
The text output in the R command line is something like
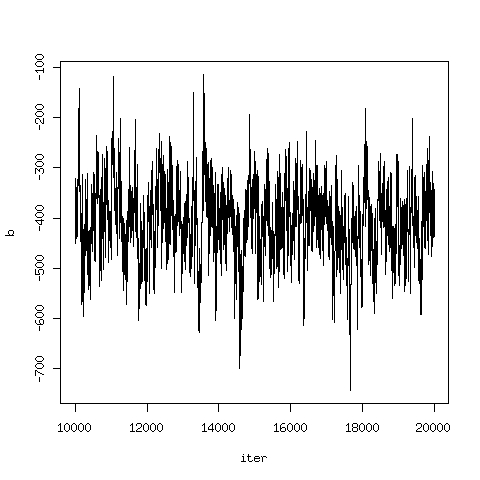
OpenBUGS ClassicBUGS release 2.1.1 type 'modelQuit()' to quit Bugs> model is syntactically correct Bugs> data loaded Bugs> model compiled Bugs> model is initialized Bugs> 10000 updates took 0 s Bugs> monitor set Bugs> monitor set Bugs> monitor set Bugs> monitor set Bugs> 10000 updates took 0 s Bugs> mean sd val2.5pc median val97.5pc sample a 8428.0 890.5 6486.0 8418.0 10220.0 10000 b -407.4 74.07 -554.9 -407.6 -243.6 10000 Bugs> mean sd MC_error val2.5pc median val97.5pc start sample a 8428.0 890.5 56.85 6753.0 8425.0 10230.0 10001 10000 b -407.4 74.07 4.753 -554.6 -406.8 -266.0 10001 10000 Bugs> CODA files written Bugs> >The output is cars which is a matrix with 10 000 rows and 3 columns. The first column is the iteration number i.e. 10 001 to 20 000 (since the burn in period was 10 000, and the chain is run for a further 10 000 iterations). The second column is the values of the parameter a for each iteration, and the third column is the equivalent for the parameter b.
You can then proceed to manipulate the cars data set e.g.
apply(cars[,-1], 2, mean)gives the mean for a to be 8427.6630 and for b it is -407.3614 which agree with the direct output from LinBugs; and
plot(cars[,c(1,3)], type="l")which gives a plot of the trajectory of the MCMC for parameter b.